Introduction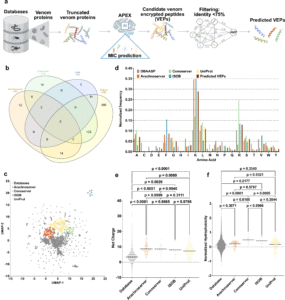
The rise of antibiotic-resistant bacteria, especially tough gram-negative strains, is one of the biggest health challenges of our time. Each year, resistant infections are responsible for around 5 million deaths worldwide, and traditional methods of discovering new antibiotics have slowed to a crawl. Venoms form an immense and largely untapped reservoir of bioactive molecules with antimicrobial potential. In the study published in Nature Communications, our team mined global venomics datasets to identify new antimicrobial candidates.
The Power of AI Meets Nature’s Deadly Defense
To explore global venomics datasets, we developed APEX, an innovative AI platform that analyzed over 16,000 venom proteins. This extensive computational process generated more than 40 million potential venom-derived peptides. From this vast pool, we identified 386 promising candidates that are both structurally and functionally distinct from known antimicrobial peptides. Of these, 58 peptides were synthesized and tested in laboratory experiments, with 53 showing effectiveness against drug-resistant bacteria. Some of these peptides also reduced bacterial loads in mice infected with Acinetobacter baumannii, a pathogen designated by the World Health Organization as a “critical priority” for new antibiotic development.
Venoms: Nature’s Evolutionary Masterpieces
Venoms from snakes, spiders, scorpions, and other creatures are among the most sophisticated chemical arsenals in nature. They have evolved over hundreds of millions of years to breach biological defenses, making them a rich, yet largely untapped, source of antimicrobial compounds. Many venom peptides disrupt cell membranes through multiple targets, making it harder for bacteria to develop resistance.
However, exploring this chemical diversity manually would take decades. That’s where AI comes into play. By pairing computational analysis with rapid laboratory validation, our team has made one of the most comprehensive investigations into venom-derived antibiotics to date. Their work has mapped over 2,000 new antibacterial motifs, vastly expanding the universe of potential peptide therapeutics.
Conclusion
reformulate This research highlights an exciting new frontier: venoms are a vast, largely untapped resource for discovering novel antibiotics. By combining large-scale computational screening with experimental validation, scientists are accelerating the development of urgently needed drugs to combat resistant infections.
For more information on this study, please refer to the full paper published in Nature Communications: https://www.nature.com/articles/s41467-025-60051-6
For more information, please contact:
Machine Biology Group
University of Pennsylvania
Authors:
Changge Guan, Marcelo D. T. Torres, Sufen Li & Cesar de la Fuente-Nunez.
Published:
July 12, 2025
About Machine Biology Group:
The mission statement of the Machine Biology Group at the University of Pennsylvania is to use the power of machines to accelerate discoveries in biology and medicine.
