
We are a transdisciplinary and diverse Team bringing together computer scientists, engineers, chemists, peptide scientists, biologists, microbiologists, and experts from various other disciplines. Our collective mission is to use the power of machines to accelerate discoveries in biology and medicine.
Explore some of our current projects:
ANTIBIOTICS AI
We developed the first antibiotic designed by a computer with proven efficacy in preclinical animal models, demonstrating that machines and AI could be used to design therapeutic molecules. This breakthrough opened new avenues for using computers for antibiotic discovery. Many of our ongoing efforts focus on the use of AI for antibiotic discovery.

MINING BIOLOGY
One of our core missions is to mine the world's biological information as a source of antibiotics and other useful molecules. Our team has been in the vanguard of developing computational methods for biological mining, leading to the discovery of a vast array of new antibiotics and significantly accelerating antibiotic discovery. We computationally mined the human proteome for the first time as a source of antibiotics. We have since mined many other proteomes, genomes, and metagenomes across the entire Tree of Life. These efforts have dramatically reduced the time needed to discover preclinical candidates, from years to hours. This work has also revealed a new class of peptides, called encrypted peptides (EPs). EPs serve as templates for antibiotic development, and we hypothesize that they may also play a role in host immunity.
MOLECULAR DE-EXTINCTION
Using machine learning, our lab has discovered the first therapeutic molecules in extinct organisms, launching the field of molecular de-extinction. We believe that resurrecting molecules from the past can help address present-day problems, such as antibiotic resistance. Indeed, resurrection biology is an emerging field that aims to bring strings of molecules and more complex organisms back to life with the ultimate goal of benefiting humanity. Thus far, we have mined the proteomes of our closest relatives, Neanderthals and Denisovans, and of all extinct organisms known to science (the "extinctome"), revealing a new universe of antimicrobial sequences, yielding preclinical candidates, and launching this emerging research area.
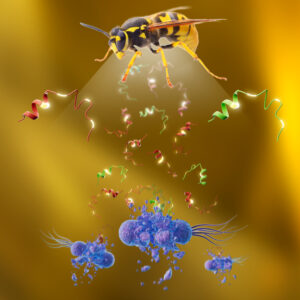 ANTIBIOTICS IN VENOMS
ANTIBIOTICS IN VENOMS
Venoms are an underappreciated source of medicines. Using structure-guided design, we have successfully reprogrammed venom-derived peptides from various creatures like wasps and frogs into potent antibiotics. These modified antibiotics have demonstrated both safety and effectiveness in preclinical mouse models. Our group has also introduced a new paradigm for treating bacterial infections through host-targeting immunotherapies, demonstrating this concept both in vitro and in animal models.


 LOW-COST DIAGNOSTICS
LOW-COST DIAGNOSTICS 