Introduction
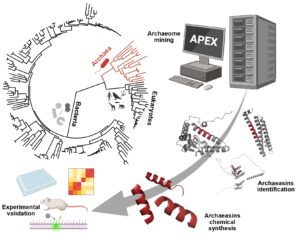 Resistance to multidrug-resistant bacteria represents one of the most serious threats to global health. Traditionally, the search for new antibiotics has focused on bacteria and fungi, which has slowed the discovery of new therapies. However, Archaea represent a virtually unexplored domain of life, with immense biochemical capabilities that could harbor previously unknown antimicrobial compounds. In this study published in Nature Microbiology, our team used deep learning to discover new antimicrobial agents within the archaeal proteome.
Resistance to multidrug-resistant bacteria represents one of the most serious threats to global health. Traditionally, the search for new antibiotics has focused on bacteria and fungi, which has slowed the discovery of new therapies. However, Archaea represent a virtually unexplored domain of life, with immense biochemical capabilities that could harbor previously unknown antimicrobial compounds. In this study published in Nature Microbiology, our team used deep learning to discover new antimicrobial agents within the archaeal proteome.
Artificial Intelligence Meets the Archaeal World
We updated our deep learning platform, APEX 1.1, to analyze 233 archaeal proteomes. This made it possible to identify 12,623 candidate molecules with potential antimicrobial activity, called archaeasins, which display amino acid patterns clearly distinct from traditional antimicrobial peptides.
From this set, 80 archaeasins were synthesized, of which 93% showed in vitro antimicrobial activity against key pathogens such as A. baumannii, E. coli, K. pneumoniae, P. aeruginosa, S. aureus, and Enterococcus spp. In in vivo evaluations using mouse models, archaeasin-73 demonstrated a bacterial load reduction capacity comparable to polymyxin B, a last-resort antibiotic.
The ability of APEX 1.1 to filter millions of potential peptides through machine learning significantly accelerated the process, providing an effective way to focus experimental development on the most promising compounds.
Archaea: An Evolutionarily Refined Source
Archaea inhabit extreme environments such as hot springs, salt flats, or hydrothermal vents, which has endowed them with unique and adaptive biochemical mechanisms. Exploring them as a source of new antimicrobials opens up a largely uncharted territory in the pharmaceutical field.
Conclusion
This research unveils a new frontier in the search for antibiotics: Archaea and their archaeasins emerge as a novel, rich source of antimicrobial molecules. By combining artificial intelligence with experimental validation, our team has opened a fast and effective path toward the development of compounds to combat bacterial resistance.
For more information on this study, please refer to the full paper published in Nature Microbiology: https://www.nature.com/articles/s41564-025-02061-0
For more information, please contact:
Machine Biology Group
University of Pennsylvania
Authors:
Marcelo D. T. Torres, Fangping Wan & Cesar de la Fuente-Nunez
Published:
August 12, 2025
About Machine Biology Group:
The mission statement of the Machine Biology Group at the University of Pennsylvania is to use the power of machines to accelerate discoveries in biology and medicine.
