Introduction
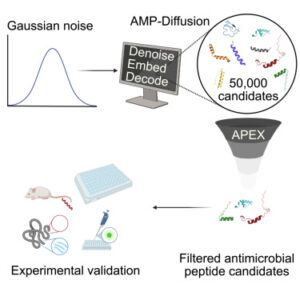 The rapid advancements in generative artificial intelligence have opened new frontiers in protein design, particularly through diffusion models that explore vast conformational and sequence spaces with unprecedented precision. Building on this momentum, our study published in Cell Biomaterials introduces AMP-Diffusion, a novel latent diffusion framework tailored for antimicrobial peptide (AMP) generation.
The rapid advancements in generative artificial intelligence have opened new frontiers in protein design, particularly through diffusion models that explore vast conformational and sequence spaces with unprecedented precision. Building on this momentum, our study published in Cell Biomaterials introduces AMP-Diffusion, a novel latent diffusion framework tailored for antimicrobial peptide (AMP) generation.
How AMP-Diffusion works
Unlike traditional approaches that require extensive protein-specific training, AMP-Diffusion directly operates within the embedding space of the powerful pre-trained language model ESM-2, harnessing its rich biological knowledge to produce functionally potent peptides. This integration allows us to generate AMPs with controlled properties, validated through comprehensive experimental assessments of their structure, activity, and therapeutic potential
From 50,000 designs to in-vivo efficacy
From an initial pool of 50,000 candidate peptides, we selected 46 based on predicted antimicrobial activity, sequence diversity, and novelty for experimental testing. The minimum inhibitory concentration (MIC) results revealed that these peptides maintained antimicrobial potency comparable to the training set, affirming the model’s ability to produce functionally relevant sequences. Additionally, naturalness assessments using the ProGen2 perplexity metric indicated that our generated peptides are as natural-looking as real AMPs, further underscoring the promise of AMP-Diffusion in designing novel antimicrobial agents with real-world potential.
Of 46 top candidates synthesized and tested experimentally, two reduced drug-resistant skin infections in mice with efficacy comparable to current clinical antibiotics, and with no observed toxicity in the studies. These findings underscore the robustness of our model in identifying potent antimicrobial peptides and highlight its potential for accelerating the development of new therapeutic agents.
Conclusion
AMP-Diffusion represents a powerful platform for antibiotic discovery, combining advanced AI techniques with experimental validation to accelerate the development of new antimicrobial agents. This approach holds promise not only for combating resistant infections but also for broadening the horizons of peptide therapeutics.
For more information on this study, please refer to the full paper published in Cell Biomaterials: https://www.cell.com/cell-biomaterials/fulltext/S3050-5623(25)00174-6
For more information, please contact:
Machine Biology Group
University of Pennsylvania
Authors:
Marcelo D. T. Torres, Leo Tianlai Chen, Fangping Wan, Pranam Chatterjee & Cesar de la Fuente-Nunez
Published:
September 2, 2025
About Machine Biology Group:
The mission statement of the Machine Biology Group at the University of Pennsylvania is to use the power of machines to accelerate discoveries in biology and medicine.
