Introduction
 Computational protein and peptide design stands out as one of the most thrilling areas of scientific advancement today, driving innovations such as new antibiotics and customized biomaterials. However, a major challenge lies in the fact that many existing design tools depend heavily on generative AI models, which demand substantial computational power and can be challenging to modify when pursuing different design objectives.
Computational protein and peptide design stands out as one of the most thrilling areas of scientific advancement today, driving innovations such as new antibiotics and customized biomaterials. However, a major challenge lies in the fact that many existing design tools depend heavily on generative AI models, which demand substantial computational power and can be challenging to modify when pursuing different design objectives.
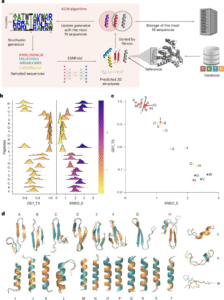
The Key-Cutting Machine (KCM) revolutionizes protein design by replacing traditional black-box generative methods with a transparent, optimization-driven approach. This system achieves high-precision outcomes efficiently, requiring only a single GPU.
From Templates to Tailored Proteins
At its core, KCM is built on a straightforward yet impactful concept: designing proteins is akin to crafting a key—tailoring a sequence to precisely match a particular backbone structure.
Instead of randomly generating sequences and hoping for optimal results, KCM employs an Estimation of Distribution Algorithm to iteratively refine amino acid sequences via structure prediction. It assesses each candidate’s adherence to geometric, physicochemical, and energetic criteria, and then uses this evaluation to guide subsequent improvements.
This optimization-driven approach allows scientists to encode user-defined objectives—like a desired structure or charge distribution—directly into the design process. That means no expensive model retraining is needed every time a new constraint is introduced.
Key Features
- Single-Device Efficiency– designs run on one GPU, opening doors for small labs and teaching settings.
- Customizable Objectives – searchers can encode specific structural or functional requirements directly into the optimization.
- Optimization-Driven – replaces black-box generation with transparent, stepwise improvement of candidate molecules.
- High Versatility – successfully designs α-helices, β-sheets, and unstructured regions with surgical accuracy.
From Algorithm to Antibiotic
By employing a well-characterized antimicrobial peptide as the “key,” KCM developed candidate compounds exhibiting strong in-vitro activity and in-vivo effectiveness. Two designs, NE2 and NG1, reduced A. baumannii bacterial burdens by up to 100-fold in mouse skin-abscess and deep-thigh infection models—achieving results comparable to polymyxin B and levofloxacin—and demonstrated favorable tolerability.
Conclusion
By combining accessibility with precision, the Key-Cutting Machine introduces a powerful new model for designing proteins de novo—no high-end hardware, no cloud credits, no AI PhD required. With applications in medicine, materials science, and synthetic biology, KCM’s accessible architecture could open new avenues in research for small labs, startups, and educators.
For more information on this study, please refer to the full paper published in Nature Machine Intelligence: https://www.nature.com/articles/s42256-025-01119-2
For more information, please contact:
Machine Biology Group
University of Pennsylvania
Authors:
Yan C. Leyva, Marcelo D. T. Torres, Carlos A. Oliva, Cesar de la Fuente-Nunez & Carlos A. Brizuela
Published:
October 21, 2025
About Machine Biology Group:
The mission statement of the Machine Biology Group at the University of Pennsylvania is to use the power of machines to accelerate discoveries in biology and medicine.
